Structural Biology
{slider-nested About}
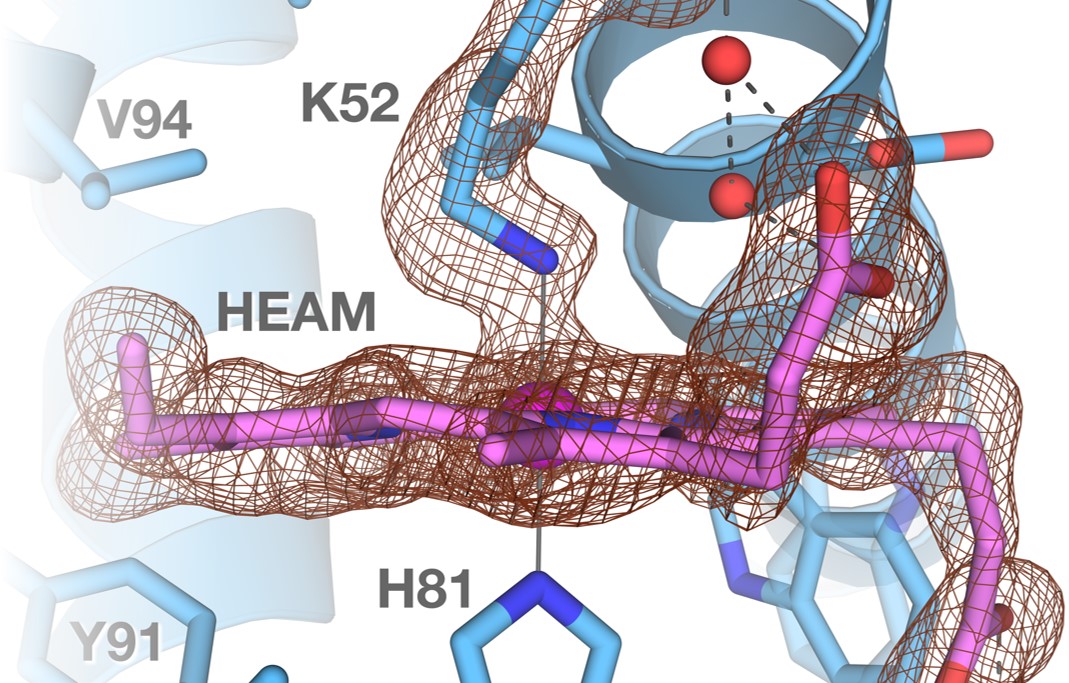
In order to understand how proteins function — for example, how they transport oxygen, detect light, fire an electrical impulse, synthesise or break down certain molecules, etc — we need to know their three-dimensional structures. Our group uses X-ray crystallography to solve the structures of proteins.
Research in our group focuses on bacterial and eukaryotic enzymes participating in metabolic pathways such as the synthesis of bioplastics, latex and fatty acids, as well as polysaccharide degradation. We are also working on signalling proteins involved in Salmonella pathogenicity and human mTOR signalling. Presently, we are collaborating with Dr Go Furusawa to study proteins from Mangrovimonas bacteria isolated by his group.
Research in our group focuses on bacterial and eukaryotic enzymes participating in metabolic pathways such as the synthesis of bioplastics, latex and fatty acids, as well as polysaccharide degradation. We are also working on signalling proteins involved in Salmonella pathogenicity and human mTOR signalling. Presently, we are collaborating with Dr Go Furusawa to study proteins from Mangrovimonas bacteria isolated by his group.

{slider-nested Research|closed}

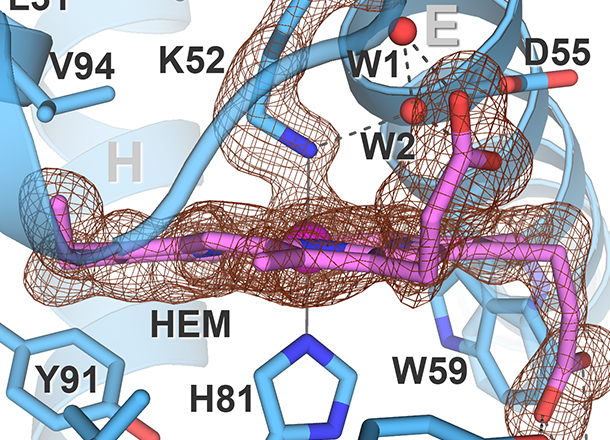
Teh, A.-H., Fazli, N.H., Furusawa, G.
Purification and characterization of new bio-plastic degrading enzyme from Burkholderia cepacia DP1
Biochemical Characterization of Thermostable and Detergent-Tolerant β-Agarase, PdAgaC, from Persicobacter sp. CCB-QB2
Crystal Structure And Epitope Analysis Of House Dust Mite Allergen Der F 21
Modelling of polyhydroxyalkanoate synthase from Aquitalea sp. USM4 suggests a novel mechanism for polymer elongation
Functional and Structural Studies of a Multidomain Alginate Lyase from Persicobacter sp. CCB-QB2
Nik Murniati Nik Man Irdahayu, Kannusamy Shantini, kai-Hee Huong, Sevakumaran Vigneswari, Nursolehah Abdul Aziz, Mohd. Noor Mohd. Azizan, Al-Ashraf Abdullah Amirul.
Microbial-based synthesis of highly elastomeric biodegradable poly(3-hydroxybutyrate-co-4-hydroxybutyrate) thermoplastic
Azami, N.A., Wirjon, I.A., Kannusamy, S., Teh, A.-H., Abdullah, A.A.-A.
Crystallization and X-ray crystallographic analysis of recombinant TylP, a putative γ-butyrolactone receptor protein from Streptomyces fradiae
Crystal Structure Of Anoxybacillus α-amylase Provides Insights Into Maltose Binding Of A New Glycosyl Hydrolase Subclass
Cloning, expression, purification, crystallization and X-ray crystallographic analysis of recombinant human c1ORF123 protein
Cloning, expression, purification, characterization, crystallization and X-ray crystallographic analysis of recombinant Der f 21 (rDer f 21) from Dermatophagoides farinae
Complete genome sequence of the thermophilic Thermus sp. CCB_US3_UF1 from a hot spring in Malaysia
Open and Lys–His hexacoordinated closed structures of a globin with swapped proximal and distal sites
Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Crystal structure of truncated hemoglobin from an extremely thermophilic and acidophilic bacterium
Crystal structure of a compact α-amylase from Geobacillus thermoleovorans

PHA Synthesis
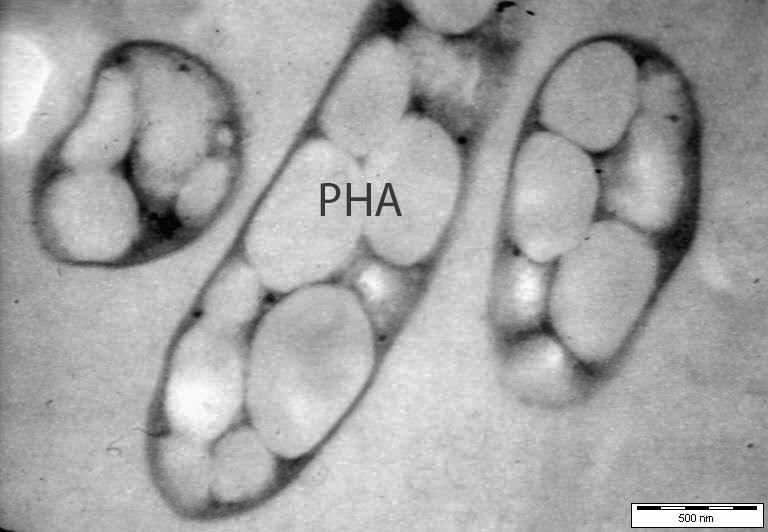
Many bacteria produce polyhydroxyalka-noates (PHAs) as carbon storage. Being biodegradable polymers, PHAs have tremendous industrial and medical uses such as bioplastics and biomaterials, and are synthesized by PHA synthases. Four classes of PHA synthases have been identified, which recognize different monomers to produce PHAs that can have extremely different physical properties. Structural information of these enzymes will not only provide insights into their exact mechanisms, but also form the basis necessary for efficiently engineering them to produce PHAs with the desired properties.
Polysaccharide Degradation
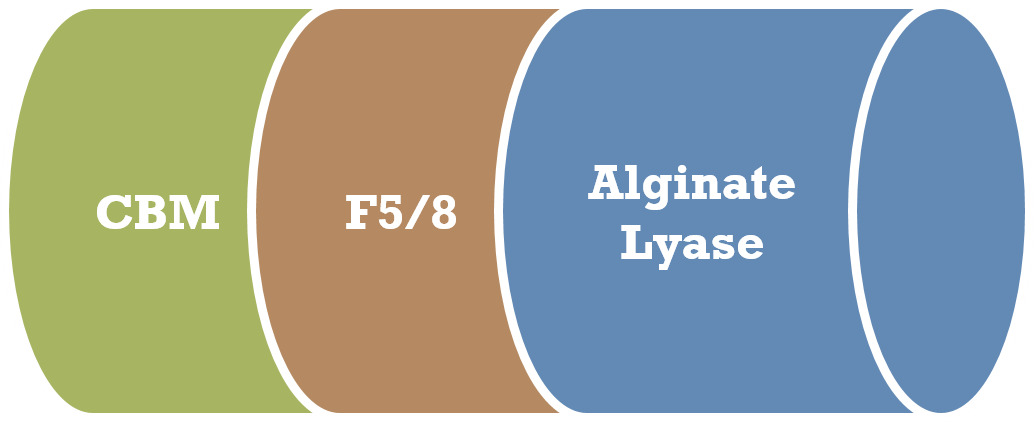
Polysaccharide degrading enzymes have biotechnological applications important to many industries such as food, paper and textile. Alginate lyases degrade an anionic polysaccharide, alginate, which is distributed in the cell walls of brown algae and can constitute up to 40% of their dry weight. Persicobacter sp., a bacterium isolated from a mangrove swamp by Dr Go Furusawa, expresses a unique alginate lyase that contains two additional carbohydrate-binding (CBM and F5/8) domains. As in recent years algal biomass has attracted worldwide attention as raw materials for biofuel production, development of efficient alginate lyases is increasingly important for the degradation of algal cell walls. We also collaborate on a bacterial agarase, whose activity is enhanced by an additional carbohydrate-binding domain.
Toxin–Antitoxin System
Toxin–antitoxin (TA) systems, composed of a toxin and an antitoxin encoded in an operon, are widespread in prokaryotes. The toxin and the antitoxin form a stable nontoxic complex in normal growing cells, but under stress the antitoxin is degraded and the toxin is released to attenuate cell growth. In the course of analyzing several Salmonella genomes, we have come across a new Type II TA system comprising a putative RNase as the toxin. This TA system is conserved not only in S. enterica, but also in pathogenic E. coli strains, Shigella dysenteriae and Klebsiella pneumoniae, indicating a possible role for the system in bacterial pathogenicity.
Fatty Acid Synthesis
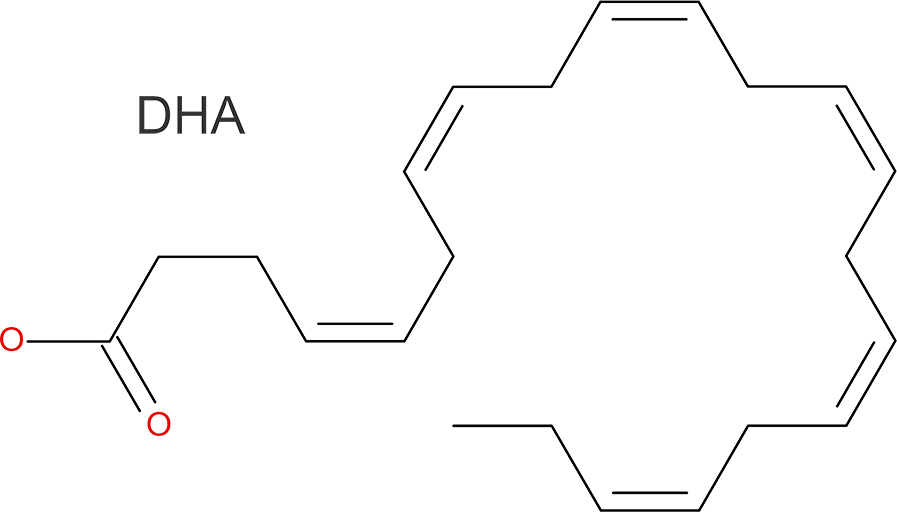
Polyunsaturated fatty acids (PUFA), including omega-3 fatty acids such as docosahexaenoic acid (DHA), are pivotal for the normal function and development of vertebrates. Fatty acid desaturases (Fads) are key enzymes in PUFA synthesis that introduce double bonds into fatty acids. In order to understand how Fads enzymes selectively act on specific sites of the long chains of PUFA, we are working on determining the structure of zebrafish Fads2, which comprises an N-terminal cytochrome b5 domain and a C-terminal desaturase domain with two transmembrane regions.
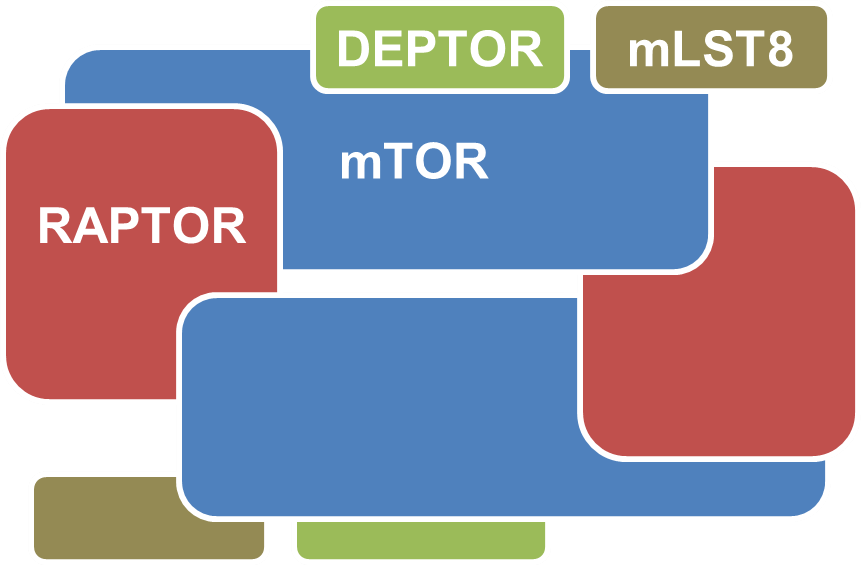
mTOR Signalling
The mTOR (mammalian target of rapamycin) signalling pathway is a central pathway that regulates cell growth and metabolism. The key protein, mTOR, is a huge serine/threonine kinase that forms two distinct complexes to regulate important kinases through phosphory-lation. The regulation of mTOR activity is critical, and deregulation of mTOR signalling has profound effects on lifespan as well as on the development of age-related diseases and cancer in human.
Globins from Extremophile
Globins are heam-containing proteins with functions like transporting, storing and sensing small molecules such as O2. The bacterium Methylacidiphilum infernorum, isolated from Hell's Gate in New Zealand, grows optimally at 60 °C and pH 2.0. Its genome encodes five globins, and we have solved the structures of three of them — HGbI, HGbIV and HGbRL. HGbI has very high affinity for O2 and is exceptionally resistant to oxidation, whereas HGbIV contains a large hydrophilic active site that can bind a phosphate ion. HGbRL, meanwhile, is a putative sensor with an additional domain, forming open and closed structures with swapped proximal and distal sites. These interesting globins most probably have gained some novel functions that help M. infernorum survive in extreme environments.{slider-nested Publications|closed}
{tab=2020}
Insights Into DEPTOR Regulation From In Silico Analysis Of DEPTOR Complexes
Teh, A.-H., Yeap, K.-H., Hisano, T.
(2020) Journal Of Structural Biology, 212 (2), Art. No. 107602,
Crystal structure of a neoagarobiose-producing GH16 family β-agarase from Persicobacter sp. CCB-QB2
Teh, A.-H., Fazli, N.H., Furusawa, G.
(2020) Applied Microbiology and Biotechnology, 104 (2), pp. 633-641.
Crystal structure of a neoagarobiose-producing GH16 family β-agarase from Persicobacter sp. CCB-QB2
Teh, A.-H., Fazli, N.H., Furusawa, G.
(2020) Applied Microbiology and Biotechnology, 104 (2), pp. 633-641.
{tab=2019}
Purification and characterization of new bio-plastic degrading enzyme from Burkholderia cepacia DP1
Azura Azami, N., Ira Aryani, W., Aik-Hong, T., Amirul, A.A.
(2019) Protein Expression and Purification, 155, pp. 35-42.
Biochemical Characterization of Thermostable and Detergent-Tolerant β-Agarase, PdAgaC, from Persicobacter sp. CCB-QB2
Hafizah, N.F., Teh, A.-H., Furusawa, G.
(2019) Applied Biochemistry and Biotechnology, 187 (3), pp. 770-781.
Crystal Structure And Epitope Analysis Of House Dust Mite Allergen Der F 21
Pang, S.L., Ho, K.L., Waterman, J., Rambo, R.P., Teh, A.-H., Mathavan, I., Harris, G., Beis, K., Say, Y.-H., Anusha, M.S., Sio, Y.Y., Chew, F.T., Ng, C.L.
(2019) Scientific Reports, 9 (1), art. no. 4933, .
{tab=2018}
Crystal structure and functional analysis of human C1ORF123
Rahaman, S.N.A., Yusop, J.M., Mohamed-Hussein, Z.-A., Aizat, W.M., Ho, K.L., Teh, A.-H., Waterman, J., Tan, B.K., Tan, H.L., Li, A.Y., Chen, E.S., Ng, C.L.
(2018) PeerJ, 2018 (9), art. no. e5377, .
Modelling of polyhydroxyalkanoate synthase from Aquitalea sp. USM4 suggests a novel mechanism for polymer elongation
Teh, A.-H., Chiam, N.-C., Furusawa, G., Sudesh, K.
(2018) International Journal of Biological Macromolecules, 119, pp. 438-445.
{tab=2017}
Functional and Structural Studies of a Multidomain Alginate Lyase from Persicobacter sp. CCB-QB2
Sim, P.-F., Furusawa, G., Teh, A.-H.
(2017), Scientific Reports (2017), 7(1),13656
En route to economical eco-friendly solvent system in enhancing sustainable recovery of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) copolymer
Nik Murniati Nik Man Irdahayu, Kannusamy Shantini, kai-Hee Huong, Sevakumaran Vigneswari, Nursolehah Abdul Aziz, Mohd. Noor Mohd. Azizan, Al-Ashraf Abdullah Amirul.
(2017), Engineering in Life Sciences, 17,1050-1059
Microbial-based synthesis of highly elastomeric biodegradable poly(3-hydroxybutyrate-co-4-hydroxybutyrate) thermoplastic
Huong K-H, Teh C.-H, Amirul A.-A.A.
(2017), International Journal of Biological Macromolecules (2017), 101:983-995
Enhanced degradation of polyhydroxyalkanoates (PHAs) by newly isolated Burkholderia cepacia DP1 with high depolymerase activity
Azami, N.A., Wirjon, I.A., Kannusamy, S., Teh, A.-H., Abdullah, A.A.-A.
(2017), 3 Biotech (2017), doi: 10.1007/s13205-017-0716-7
Crystallization and X-ray crystallographic analysis of recombinant TylP, a putative γ-butyrolactone receptor protein from Streptomyces fradiae
Mohd-Sharif N., Shaibullah S., Baharum S.N., Ng C.L., Givajothi V., Tan C.-S., Ho K.L., Teh A.-H., Waterman J.
(2017), Acta Cryst. F , 73,109-115
{tab=2016}
Crystal Structure Of Anoxybacillus α-amylase Provides Insights Into Maltose Binding Of A New Glycosyl Hydrolase Subclass
Kian Piaw Chai, Noor Farhan Binti Othman. Aik-Hong Teh, Kok Lian Ho, Kok-Gan Chan, Mohd Shahir Shamsir, Kian Mau Goh & Chyan Leong Ng.
(2016), Scientific Report, 6, Article number:23126.
Cloning, expression, purification, crystallization and X-ray crystallographic analysis of recombinant human c1ORF123 protein
Rahaman, S.N.A, Yusop, J.M., Mohamed-Hussein, Z.-A, Ho, K.L., Teh, A.-H, Waterman, J., Ng, C.L..
(2016), Acta Cryst. F, 72, 207-213.
{tab=2015}
Cloning, expression, purification, characterization, crystallization and X-ray crystallographic analysis of recombinant Der f 21 (rDer f 21) from Dermatophagoides farinae
S. L. Pang, K. L. Ho, J. Waterman, A.-H. Teh, F. T. Chew and C. L. Ng.
(2015), Acta Crystallographica Section F: Structural Biology Communications 71, 1396–1400 (2015).
Complete genome sequence of the thermophilic Thermus sp. CCB_US3_UF1 from a hot spring in Malaysia
Beng Soon Teh, Nyok-Sean Lau, Fui Ling Ng, Ahmad Yamin Abdul Rahman, Xuehua Wan, Jennifer A. Saito, Shaobin Hou, Aik-Hong Teh, Nazalan Najimudin, and Maqsudul Alam.
(2015), Standards in Genomic Sciences, , 10(2015), 76
Open and Lys–His hexacoordinated closed structures of a globin with swapped proximal and distal sites
Teh AH, Saito JA, Najimudin N, Alam M.
(2015), Scientific Reports 5, 11407 (2015).
Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Teh AH, Makino M, Hoshino T, Baba S, Shimizu N, Yamamoto M, Kumasaka T.
(2015), Acta Crystallographica Section D: Biological Crystallography 71, 1392–1399 (2015).
{tab=2014}
Crystal structure of truncated hemoglobin from an extremely thermophilic and acidophilic bacterium
Farrukh Jamil,*, Aik-Hong Teh, Ermin Schadich, Jennifer A. Saito, Nazalan Najimudin, and Maqsudul Alam.
(2014), J Biochem (2014) doi: 10.1093/jb/mvu023
{tab=2013}
Crystal structure of a compact α-amylase from Geobacillus thermoleovorans
Mok S.-C., Teh A.-H., Saito J.A., Najimudin N., Alam M.
(2013), Enzyme and Microbial Technology, 53,46-54
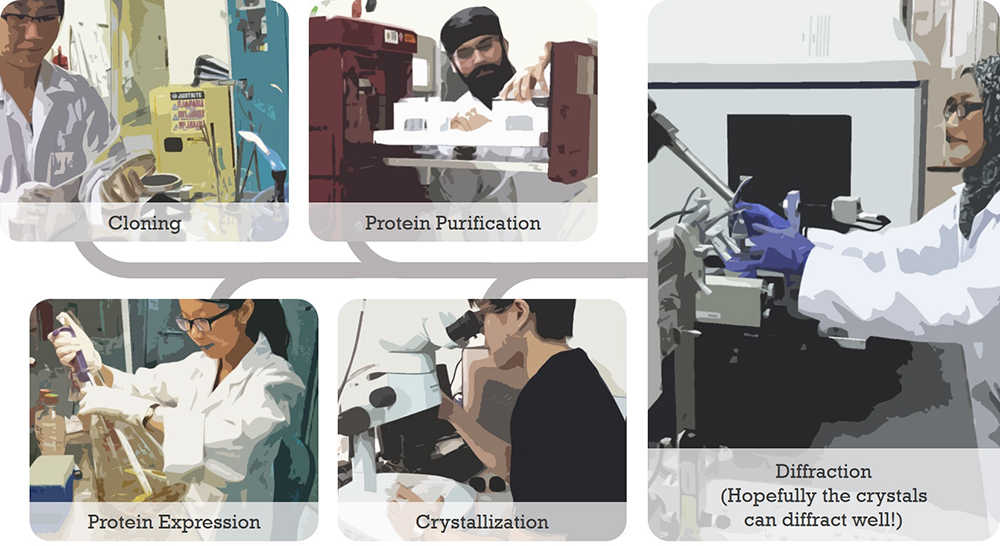
{slider-nested Protein Crystallography at a Glance|closed}

{slider-nested Members|closed}
{slider-nested Services|closed}
We also offer services for protein X-ray crystallography.
{/sliders-nested}
- Hits: 11217