Environmental Microbiology
|
 |
|
|
|
|
|
{tab=2020}
Crystal structure of a neoagarobiose-producing GH16 family β-agarase from Persicobacter sp. CCB-QB2
Teh, A.-H., Fazli, N.H., Furusawa, G.
(2020) Applied Microbiology and Biotechnology, 104 (2), pp. 633-641.
{tab=2019}
Complete Genome Sequence Of The Novel Agarolytic Catenovulum-Like Strain CCB-QB4
Lau, N.-S., Tan, W.R., Furusawa, G., Amirul, A.-A.A.
(2019) Marine Genomics, 43, pp. 50-53.
Biochemical Characterization of Thermostable and Detergent-Tolerant β-Agarase, PdAgaC, from Persicobacter sp. CCB-QB2
Hafizah, N.F., Teh, A.-H., Furusawa, G.
(2019) Applied Biochemistry and Biotechnology, 187 (3), pp. 770-781.
Draft genome sequence of the halophilic Pararhodobacter-like strain CCB-MM2, which has polyhydroxyalkanoate-synthesizing potential
Sam, K.-K., Lau, N.-S., Furusawa, G., Abdullah Amirul, A.-A.
(2019) Microbiology Resource Announcements, 8 (46), art. no. e01248-19,
Biodiversity of plant polysaccharide-degrading bacteria in mangrove ecosystem
Furusawa, G.
(2019) Tropical Life Sciences Research, 30 (3), pp. 157-172.
{tab=2018}
Metagenomic insights into the phylogenetic and functional profiles of soil microbiome from a managed mangrove in Malaysia
Priya, G., Lau, N.-S., Furusawa, G., Dinesh, B., Foong, S.Y., Amirul, A.-A.A.
(2018) Agri Gene, 9, pp. 5-15.
Modelling of polyhydroxyalkanoate synthase from Aquitalea sp. USM4 suggests a novel mechanism for polymer elongation
Teh, A.-H., Chiam, N.-C., Furusawa, G., Sudesh, K.
(2018) International Journal of Biological Macromolecules, 119, pp. 438-445.
{tab=2017}
Functional and Structural Studies of a Multidomain Alginate Lyase from Persicobacter sp. CCB-QB2
Sim, P.-F., Furusawa, G., Teh, A.-H.
(2017), Scientific Reports, 7(1),13656
Draft genome sequence of halophilic Hahella sp. strain CCB-MM4, isolated from Matang Mangrove Forest in Perak, Malaysia
Ka-Kei Sam, Nyok Sean Lau, Go Furusawa, Al-Ashraf Abdullah Amirul.
(2017), Genome Announcements, 5(42),e01147-17
Microbulbifer aggregans sp. nov., isolated from estuarine sediment from a mangrove forest
Moh, T.H., Furusawa, G., Amirul, A.A.-A.
(2017), nternational Journal of Systematic and Evolutionary Microbiology (2017), 67(10),002258, pp. 4089-4094.
Complete genome sequence of Microbulbifer sp. CCB-MM1, a halophile isolated from Matang Mangrove Forest, Malaysia
Moh, T.H., Lau, N.-S., Furusawa, G., Amirul, A.-A.A.
(2017), Standards in Genomic Sciences (2017), 12(1),36
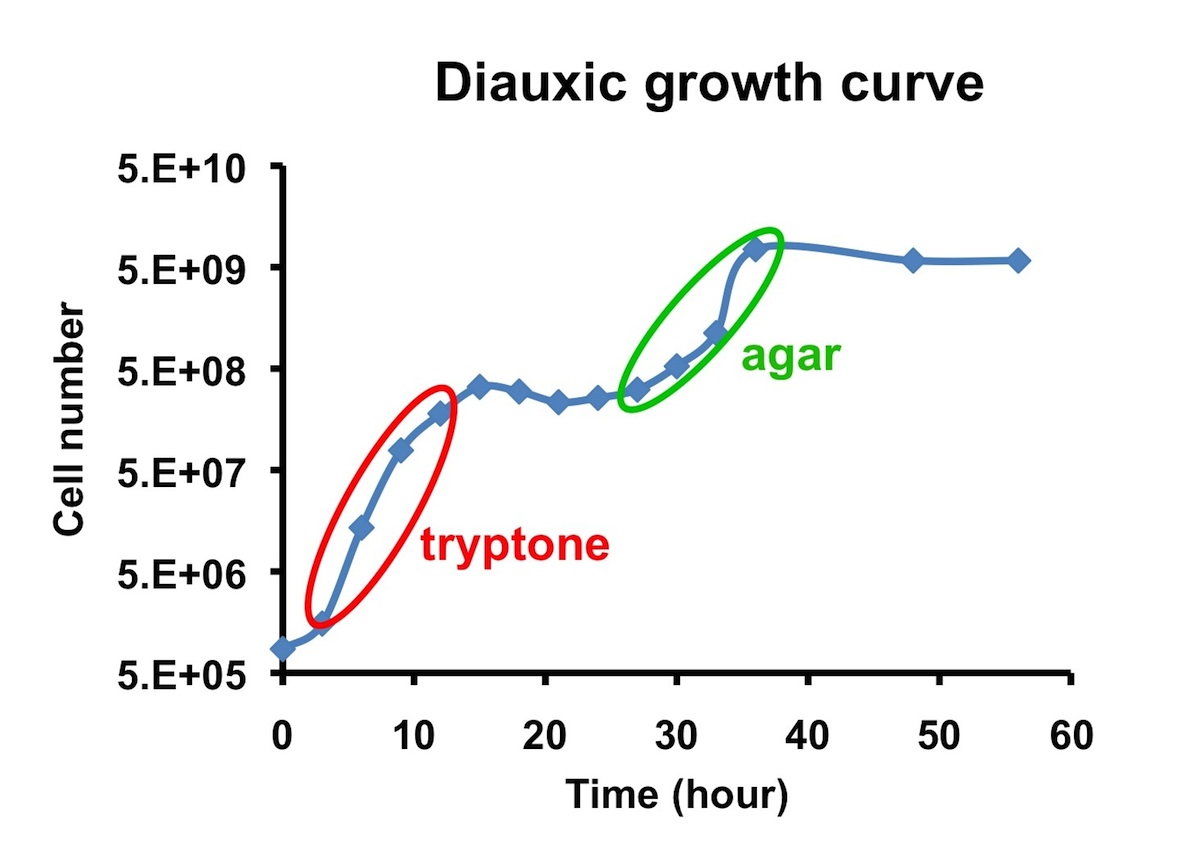
Agarolytic bacterium Persicobacter sp. CCB-QB2 exhibited a diauxic growth involving galactose utilization pathway
Go Furusawa, Nyok-Sean Lau, Suganthi Appalasamy, Amirul Al-Ashraf Abdullah.
(2017), Microbiology Open, doi: 10.1002/mbo3.405
Mangrovimonas xylaniphaga sp. nov. isolated from estuarine mangrove sediment of matang mangrove forest, malaysia
Balachandra Dinesh, Go Furusawa, A. A. Amirul. 2017.
(2017), Archives of Microbiology, 199-63-67
{tab=2016}
Comparative genome analyses of novel Mangrovimonas-like strains isolated from estuarine mangrove sediments reveal xylan and arabinan utilization genes
Balachandra Dinesh, Nyok-Sean Lau, Go Furusawa, Seok-Won Kim, Todd D. Taylor, Swee Yeok Foong, Alexander Chong Shu-Chien.
(2016), Marine Genomics, 25 (2016), 115-121.
Mangrovimonas xylaniphaga sp. nov. isolated from estuarine mangrove sediment of Matang Mangrove Forest, Malaysia
Dinesh B., Furusawa G., Amirul A.A.
(2016), Archives of Microbiology, 1-5
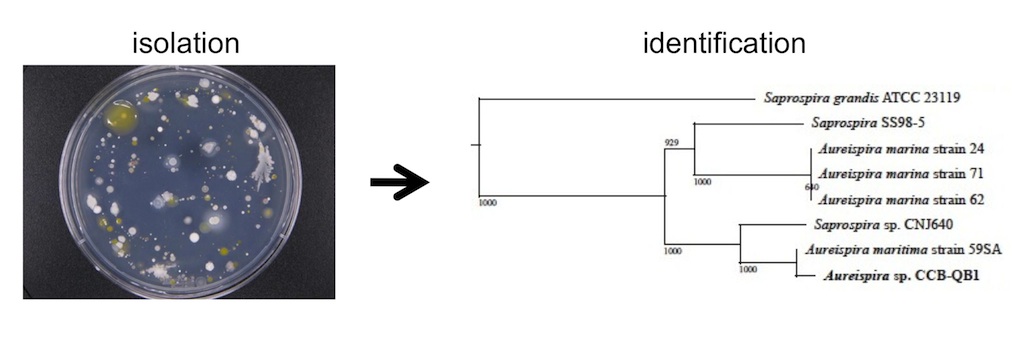
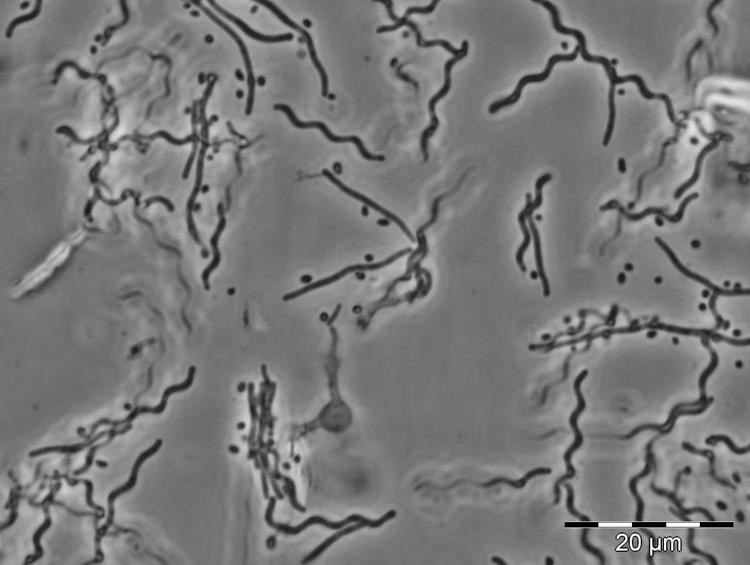
Calcium is required for ixotrophy of Aureispira sp. CCB-QB1
Furusawa, G., Hartzell, P. L., & Navaratnam, V.
(2015), Microbiology , 161(10), 1933-1941
Identification of polyunsaturated fatty acid and diterpenoid biosynthesis pathways from draft genome of Aureispira sp. CCB-QB1
Furusawa G, Lau NS, Shu-Chien AC, Jaya-Ram A, Al-Ashraf AA.
(2015), Marine Genomics, 19 (2015), 39-44.
{slider-nested Members|closed}
- Hits: 4580